
咨询服务电话:400 8717 699

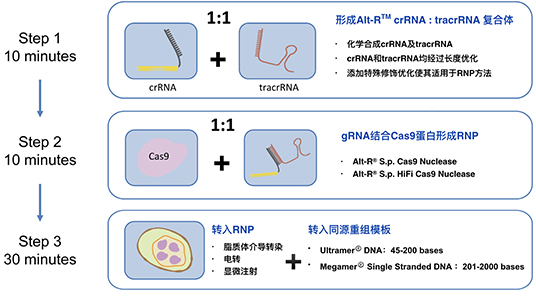
1小时3步组装具有编辑功能的Alt-R® RNP
多种方式高效导入Alt-R® RNP
提供针对不同模式生物的实验方法
Alt-R® RNP可在体外对DNA进行编辑
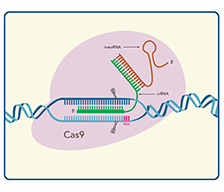
crRNA: tracrRNA 优化长度显著改善编辑效率
特殊修饰crRNA:tracrRNA 增加核酸酶抗性降低免疫反应
多物种在线crRNA预设计数据库及检查器
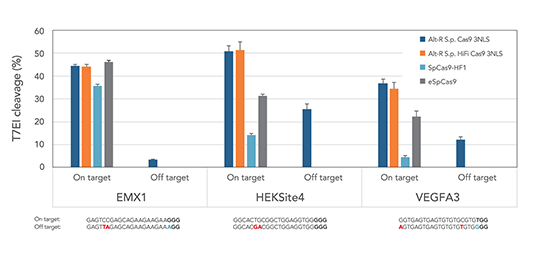
对目标位点较高的编辑效率
基因编辑脱靶效应显著降低
与gRNA复合体形成RNP,细胞毒性低,不易引起免疫反应









上海纳昂达生物科技有限公司 | Integrated DNA Technologies中国大陆授权代理商
Add:上海市杨浦区政立路477号同和国际大厦A座605室
Tel:400 8717 699 | sales@nanodigmbio.com | www.nanodigmbio.com
